AlvaDesc is the next-generation tool designed for the seamless calculation of a wide range of molecular descriptors, molecular fingerprints, and user-defined structural patterns. Featuring a simple, intuitive user interface, alvaDesc makes it easy to calculate and analyze molecular data.
Whether you’re working with molecular descriptors, identifying specific structural patterns, or generating molecular fingerprints, alvaDesc provides powerful tools for in-depth cheminformatics analysis (Mauri, A. (2020). alvaDesc: A Tool to Calculate and Analyze Molecular Descriptors and Fingerprints. In K. Roy (Ed.), Ecotoxicological QSARs (pp. 801–820). Humana Press Inc. https://doi.org/10.1007/978-1-0716-0150-1_32; Mauri, A., & Bertola, M. (2022). Alvascience: A New Software Suite for the QSAR Workflow Applied to the Blood–Brain Barrier Permeability. International Journal of Molecular Sciences, 23(21), 12882. https://doi.org/10.3390/ijms232112882).
Molecular Descriptors
It calculates almost 6,000 descriptors, among them more than 4,000 descriptors are independent of 3-dimensional information such as constitutional, topological, pharmacophore. It includes ETA and Atom-type E-state indices together with functional groups and fragment counts. Additionally, alvaDesc implements an extensive number of 3-dimensional descriptors such as 3D-autocorrelation, Weighted Holistic Invariant Molecular descriptors (WHIM), GETAWAY and Solvent Accessible Surface Area descriptors.
If needed, alvaDesc can calculate partial charges using the Gasteiger’s “Partial Equalization of Orbital Electronegativity” (PEOE) and 3D coordinates using Distance Geometry (DG). The initially generated coordinates are then refined through an optimization phase based on Universal Force Field (UFF).
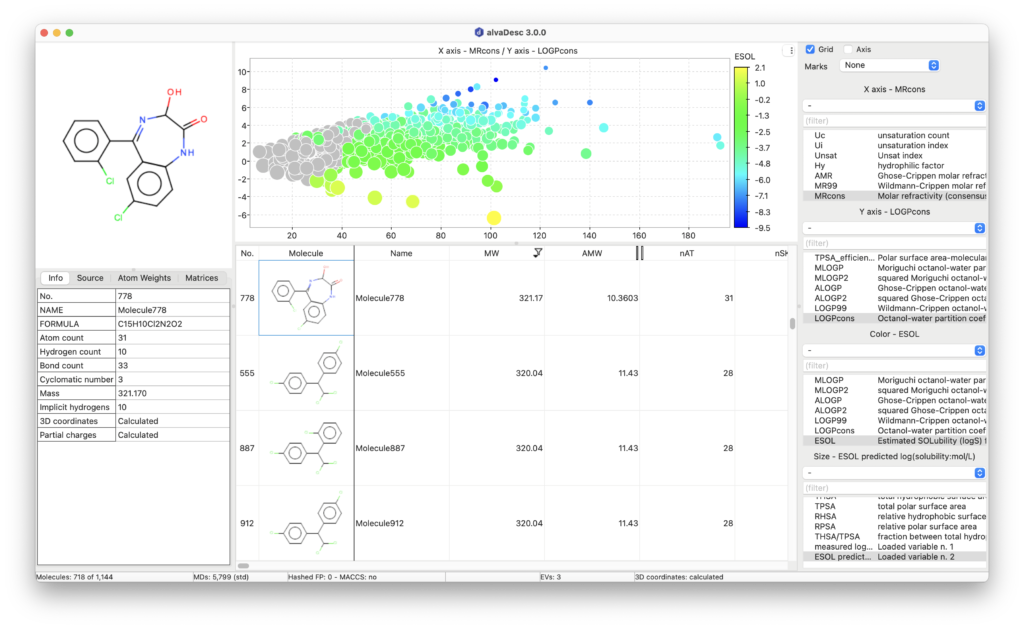
Molecular Properties, Drug-like and Lead-like Indices
AlvaDesc provides the calculation of several model-based physicochemical properties such as molar refractivity, topological polar surface area (TPSA), molecular volume estimations, three LogP models (Moriguchi, Ghose-Chippen and Wildmann-Crippen octanol-water partition coefficient), a LogP consensus model (LOGPcons) and a LogS aqueous solubility model (ESOL).
In order to get a quantitative estimation of synthetic accessibility of molecules, alvaDesc includes the synthetic accessibility score of drug-like molecules (SAscore).
There is a significative list of drug-like and lead-like alerts including the well-known Lipinski alert index. Considering the importance of drug-likeness when selecting compounds in the early stages of drug discovery, alvaDesc includes the calculation of the quantitative estimate of drug-likeness (QED).
Molecular Fingerprints
AlvaDesc carries out the calculation of MACCS166 fingerprint, Extended Connectivity Fingerprint (ECFP and ECFPV3) and Path Fingerprint (PFP) and allows the customisation and calculation of the most used hashed molecular fingerprints. The calculation of hashed fingerprints can be tuned not only with respect to the fingerprint size, fragment type and dimensions but even by defining atom and bond parameters considered during fragment identifications (e.g., atom type, aromaticity, the number of attached hydrogen atoms, connectivity).
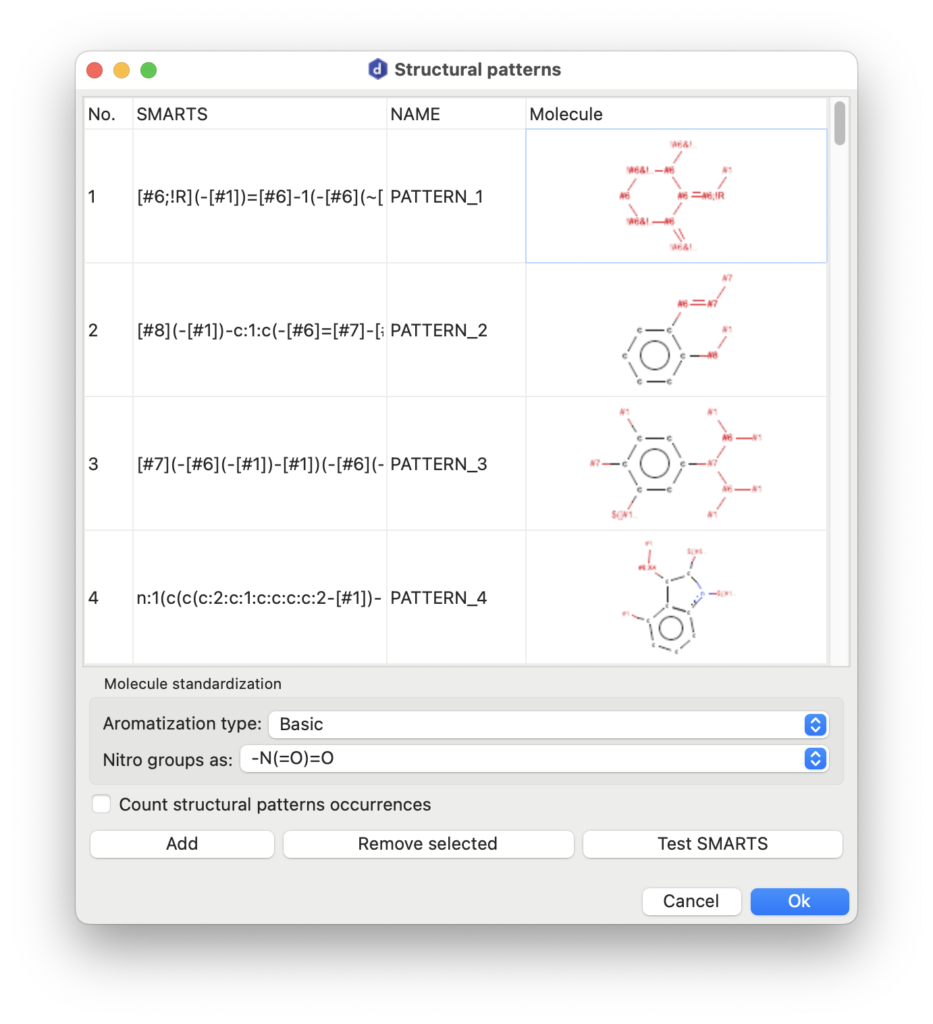
Structural Patterns
AlvaDesc can identify structural pattern occurrences in molecular datasets. A structural pattern refers to a specific arrangement of atoms and bonds within a molecule, identifiable and characterizable as a substructure. These patterns represent recurring molecular features, such as functional groups or distinct atom connectivities. Structural patterns play a pivotal role in cheminformatics, particularly in structure-activity relationship (SAR) studies, pattern recognition, and the prediction of molecular properties and behavior.
In alvaDesc, structural patterns are defined using the SMARTS syntax.
Other Features
One of the most innovative features of alvaDesc is its capability to handle both full-connected and non-full-connected molecular structures, such as salts and ionic liquids. All of the molecular descriptor calculation algorithms provide different theoretical approaches for the calculation of molecular descriptors on such structures.
Different tools are provided to carry out a first exploration of your molecular dataset:
- Molecule structure verification using PubChem and Google Patents services
- Molecule structure visualisation and filtering
- Principal Component Analysis (PCA), t-SNE and correlation analysis
Due to its capability of calculating large numbers of molecular descriptors, alvaDesc provides variable reduction tools, including the fast V-WSP method (variable reduction method adapted from space-filling designs).
Video
A short video introduction:
Platforms
The software is 64bit and it’s available for Windows, Linux and macOS. It is provided both as an easy to use command line tool and as an intuitive graphical interface.
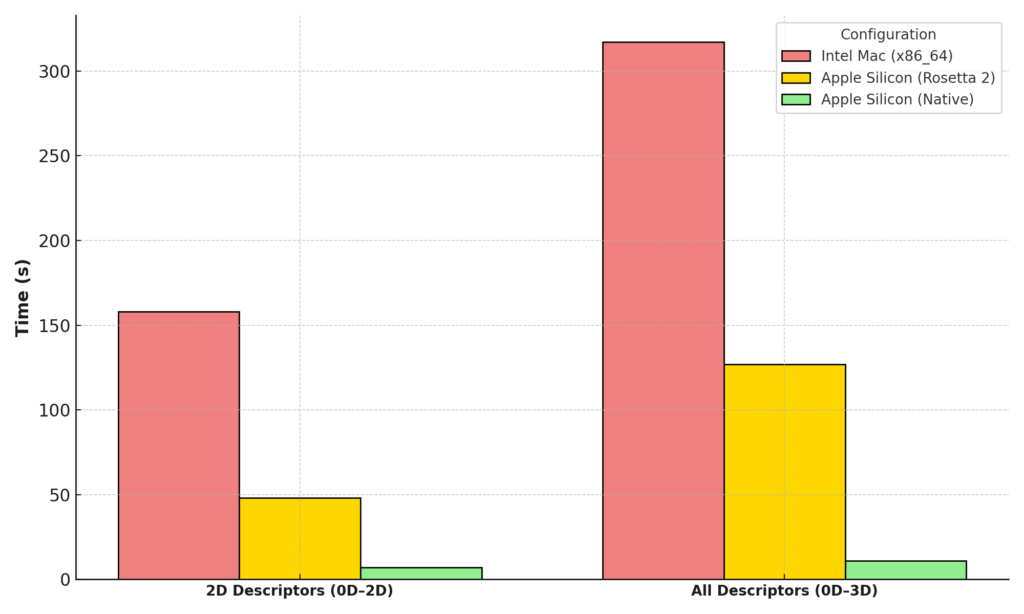
With the release of alvaDesc 3.0, experience significantly enhanced calculation speeds on M Series processors. Learn more about the benchmarking results and performance improvements here.
How to Cite
If you reference alvaDesc in an academic paper or publication, you can find the correct citation for your version by:
- Running alvaDescGUI and selecting “About alvaDesc” from the menu
- Using the command alvaDescCLI –cite
Additionally, please consider citing the following papers:
- Mauri, A. (2020). alvaDesc: A Tool to Calculate and Analyze Molecular Descriptors and Fingerprints. In K. Roy (Ed.), Ecotoxicological QSARs (pp. 801–820). Humana Press Inc. https://doi.org/10.1007/978-1-0716-0150-1_32
- Mauri, A., & Bertola, M. (2022). Alvascience: A New Software Suite for the QSAR Workflow Applied to the Blood–Brain Barrier Permeability. International Journal of Molecular Sciences, 23(21), 12882. https://doi.org/10.3390/ijms232112882
Related tools
- The perfect tool to prepare your molecular dataset for alvaDesc is alvaMolecule
- Create QSAR/QSPR models with alvaModel starting from an alvaDesc project
- A tutorial showing how to build a QSAR model using Alvascience tools
AlvaDesc is also available as:
- A set of KNIME nodes. Please check the alvaDesc KNIME Plugin page for more information.
- A Python module. Please check the alvaDescCLIWrapper page for more information.

