
With the release of alvaDesc 3.0, all Alvascience tools, including alvaDesc, alvaBuilder, alvaModel, alvaRunner, and alvaMolecule, are now fully optimized to run natively on Apple Silicon (M Series chips). These tools are distributed as Universal Binary, ensuring compatibility across both Intel-based Macs (x86_64) and Apple Silicon-based Macs (M1, M2, M1 Max, M1 Ultra, etc.) without relying on emulation layers.
Native Apple Silicon Support
Previously, Alvascience applications on Apple Silicon required the use of Rosetta 2, Apple’s emulation layer that allows x86_64 applications to run on ARM-based hardware. While functional, this approach introduced performance overheads, especially for computationally intensive tasks like descriptor calculation.
The transition to native support ensures that Alvascience tools now leverage the full computational power of Apple Silicon hardware, resulting in substantial performance gains.
Performance evaluation
To assess the impact of native Apple Silicon builds, we conducted benchmark tests using alvaDesc 3.0 and the AqSolDB dataset (Sorkun et al., 2019), which includes:
- 9,982 molecules
- 33 average atoms per molecule (maximum 670)
- 266.70 average molecular weight (maximum 5,299.8)
- 2.4 average molecular circuits (maximum 264)
We evaluated descriptor calculation times for:
- 2D Descriptor: including 0D, 1D, and 2D descriptors (4,215 descriptors)
- All descriptors: including 0D, 1D, 2D, and 3D descriptors (5,799 descriptors)
Testing Environments
We tested alvaDesc 3.0 using alvaDescCLI 3.0.4 on the following systems:
- Intel Mac: 2 GHz Quad-Core Intel Core i5, 16 GB RAM
- Apple Silicon Mac: Apple M2 Pro, 16 GB RAM
For comparison, an Intel x86 system was included in the benchmarks; however, the key takeaway is the substantial performance improvement achieved by running the native executable on Apple Silicon, which significantly reduces computation time.
Benchmark Results
The benchmark tests were conducted to evaluate the performance of alvaDesc across different system configurations, focusing on descriptor calculation times for both 2D and all descriptors (including 3D).
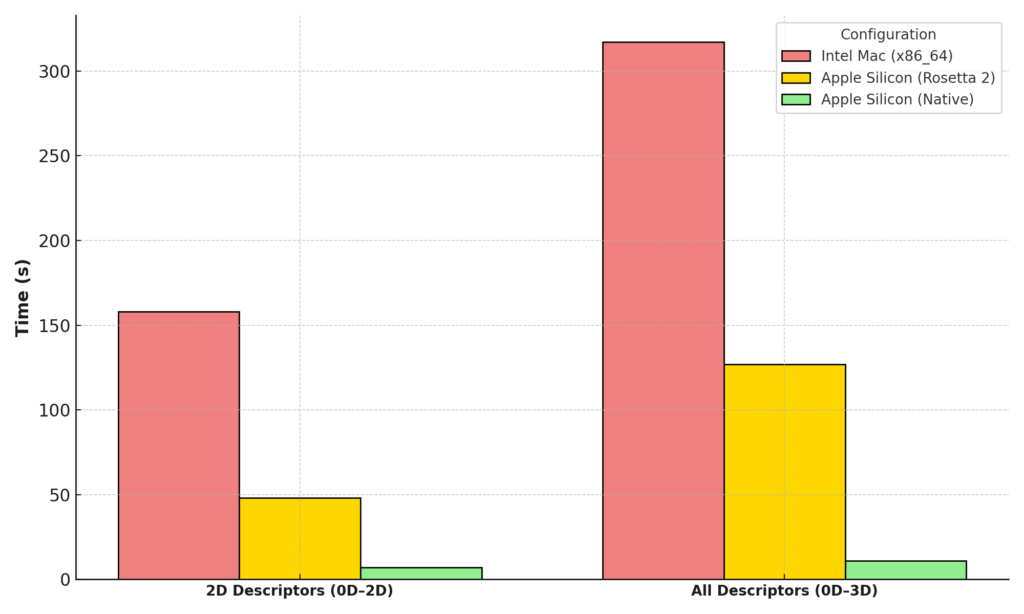
Performance comparison of descriptor calculation times for 2D (4,215 descriptors) and all descriptors (5,799 descriptors, including 3D) across three configurations: Intel Mac (x86_64), Apple Silicon via Rosetta 2, and native Apple Silicon. Native Apple Silicon builds significantly reduce computation time, demonstrating the benefits of optimized support for M Series processors.
| 2D Descriptors (0D-2D) | All Descriptors (0D–3D) | |||
|---|---|---|---|---|
| Configuration | Time | Avg. Time/Molecule (ms) | Time | Avg. Time/Molecule (ms) |
| x86_64 on Intel Mac | 2 min 38 sec | 16 | 5 min 17 sec | 32 |
| x86_64 on Apple Silicon (Rosetta 2) | 48 sec | 5 | 2 min 7 sec | 13 |
| Native Apple Silicon (M2 Pro) | 7 sec | 0.7 | 11 sec | 1.1 |
Performance comparison of descriptor calculation times across different configurations (Intel Mac, Apple Silicon via Rosetta 2, and native Apple Silicon). The table includes total calculation times for 2D descriptors (4,215 descriptors) and all descriptors (5,799 descriptors) along with average calculation time per molecule.
Technical Features of Universal Binary Distribution
- Seamless Compatibility: A single binary supports both Intel and Apple Silicon architectures, simplifying deployment.
- Optimized Performance: By leveraging Apple Silicon’s ARM architecture, computational tasks like descriptor generation and QSAR modeling are executed with enhanced efficiency.
- Improved User Experience: Faster calculations result in reduced waiting times, enabling users to process larger datasets effortlessly.
References
- Sorkun, M. C., Khetan, A., & Er, S. (2019). AqSolDB, a curated reference set of aqueous solubility and 2D descriptors for a diverse set of compounds. Scientific Data, 6(1), 143. https://doi.org/10.1038/s41597-019-0151-1
- Mauri, A. (2020). alvaDesc: A tool to calculate and analyze molecular descriptors and fingerprints. In Methods in Pharmacology and Toxicology. https://doi.org/10.1007/978-1-0716-0150-1_32
- Mauri, A., & Bertola, M. (2022). Alvascience: A New Software Suite for the QSAR Workflow Applied to the Blood–Brain Barrier Permeability. International Journal of Molecular Sciences, 23(21). https://doi.org/10.3390/ijms232112882
- Mauri, A., & Bertola, M. (2024). AlvaBuilder: A Software for De Novo Molecular Design. Journal of Chemical Information and Modeling, 64(7), 2136–2142. https://doi.org/10.1021/acs.jcim.3c00610
