In the papers “Cassotti, M., Ballabio, D., Consonni, V., Mauri, A., Tetko, I. V., & Todeschini, R. (2014). Prediction of Acute Aquatic Toxicity toward Daphnia Magna by using the GA- k NN Method. Alternatives to Laboratory Animals, 42(1), 31–41. https://doi.org/10.1177/026119291404200106” and “Cassotti, M., Consonni, V., Mauri, A., & Ballabio, D. (2014). Validation and extension of a similarity-based approach for prediction of acute aquatic toxicity towards Daphnia magna. SAR and QSAR in Environmental Research, 25(12), 1013–1036. https://doi.org/10.1080/1062936X.2014.977818“, the authors presented QSAR models to predict acute aquatic toxicity (LC50 48 hours) towards Daphnia magna.
The information described in the papers were used to build the alvaRunner project that we present here. The models were created using the first paper’s dataset consisting of 546 organic molecules.
alvaRunner project
This alvaRunner project contains four regression models:
- KNN_MD_Training: a KNN based on molecular descriptors (MD) built using the 436 molecules of the paper training set
- KNN_MD_All: a KNN based on molecular descriptors built using all the 546 molecules
- KNN_ECFP_All: a KNN based on extended connectivity fingerprints (ECFP) built using all the molecules
- Consensus_All: a Consensus using KNN_MD_All and KNN_ECFP_All
The first two models include the following eight molecular descriptors:
- MLOGP: Moriguchi octanol-water partition coefficient
- RDCHI: reciprocal distance sum Randic-like index
- SAacc: surface area of acceptor atoms from P_VSA-like descriptors
- TPSA(tot): topological polar surface area using N,O,S,P polar contributions
- H-050: H attached to heteroatom
- nN: number of Nitrogen atoms
- C-040: number of carbon atoms of type R-C(=X)-X, R-C≡X, X=C=X
- GATS1p: Geary autocorrelation of lag 1 weighted by polarizability
Cassotti highlighted that RDCHI encodes information about molecular size and branching and can be associated to lipophilicity, SAacc and TPSA(tot) account for the exposed molecular polar surface area that can interact with biological targets, H-050 contains information related to the possibility of H-bond formation, nN encodes information on the nucleophilicity, deriving from the presence of nitrogen atoms in the toxicants. C-040 seems to account for electrophilic features and GATS1p encodes information on molecular polarisability.
It’s worth noticing that the papers’ KNN models use a weighting formula that is slightly different from the one used by alvaModel/alvaRunner. Also, the paper presents an Applicability Domain using ad hoc distance thresholds. Due to the specificity of this solution, we decided not to include an Applicability Domain in this project.
The scores of the models of the alvaRunner project are presented in the following table:
| Model name | Training | Test | ||||
|---|---|---|---|---|---|---|
| R2 | Q2CV | RMSE | RMSECV | R2 | RMSE | |
| KNN_MD_Training (k: 3, Mahalanobis) | 0.595 | 0.602 | 1.059 | 1.049 | 0.43 | 1.258 |
| KNN_MD_All (k: 5, Mahalanobis) | 0.591 | 0.568 | 1.064 | 1.093 | – | – |
| KNN_ECFP_All (k: 5, Jaccard / Tanimoto) | 0.606 | 0.57 | 1.045 | 1.091 | – | – |
| Consensus_All | 0.65 | 0.626 | 0.984 | 1.017 | – | – |
The following charts show the predicted (Y) and real (X) values of the models:
| KNN_MD_Training | KNN_MD_All |
|---|---|
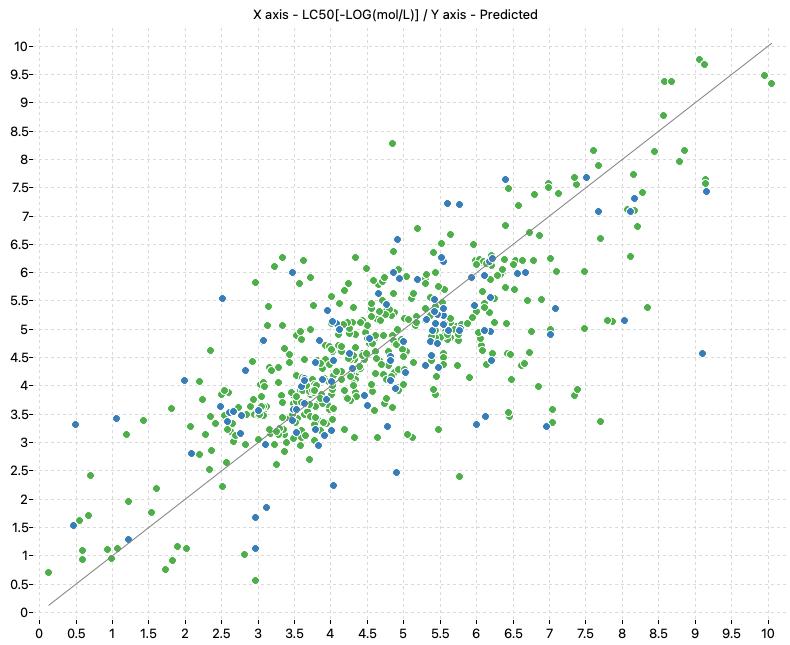 |
 |
| KNN_ECFP_All | Consensus_All |
|---|---|
 |
 |
