Model studies
QSAR and QSPR models

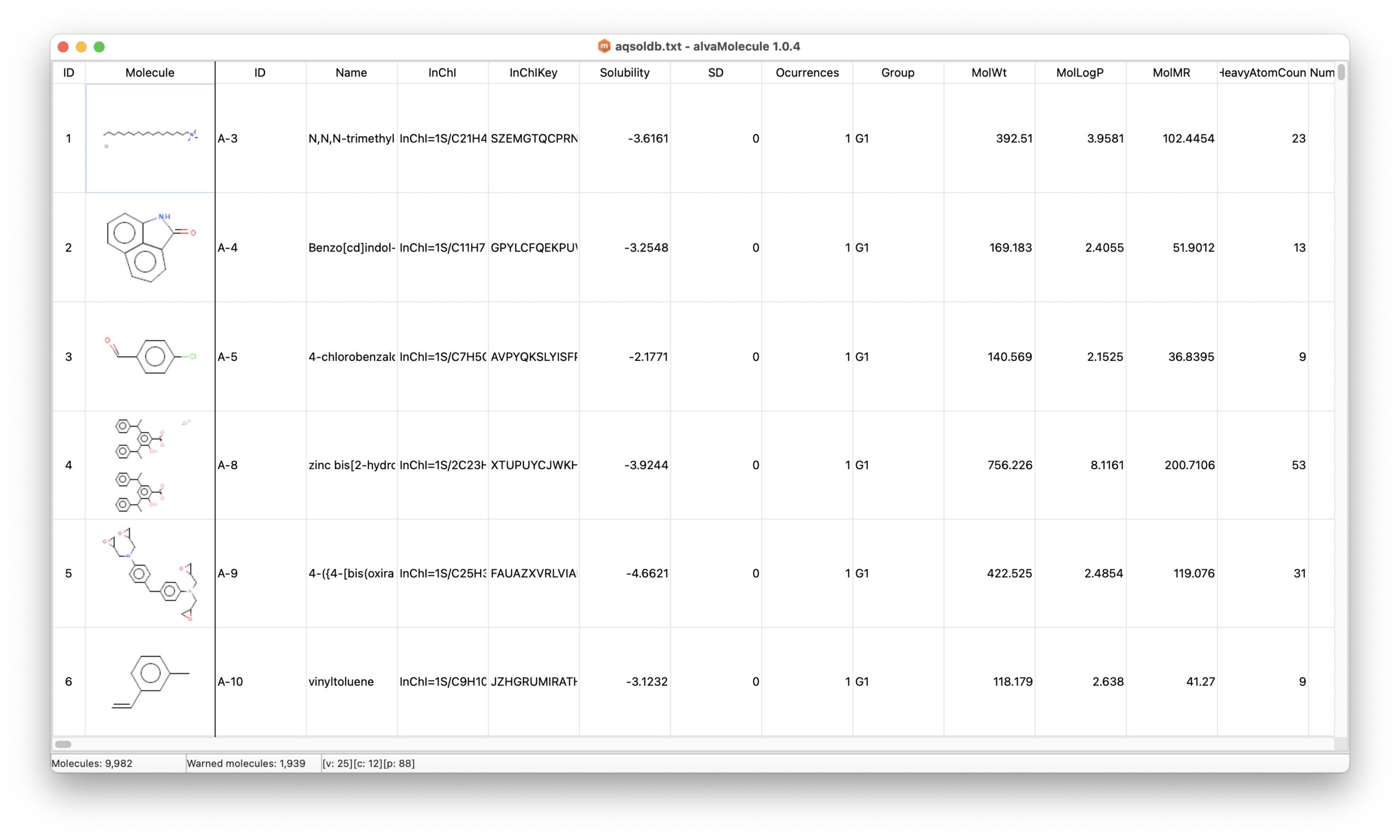
Tutorial: models for aqueous solubility (LogS)
This tutorial describes how to build a QSPR model using Alvascience tools. The selected endpoint is the aqueous solubility (LogS). The scope of this tutorial is to describe, step-by-step, the development of QSAR/QSPR regression models, from data curation to the use of models with a new set of molecules.
Prediction of the fish Biomagnification Factor (BMF) of chemicals
This project includes the models presented in the paper “Grisoni, F., Consonni, V., & Vighi, M. (2018). Acceptable-by-design QSARs to predict the dietary biomagnification of organic chemicals in fish. Integrated Environmental Assessment and Management, 15(1), 51–63. https://doi.org/10.1002/ieam.4106“. The authors presented QSAR regression models to predict the laboratory-based fish Biomagnification Factor (BMF) of chemicals.
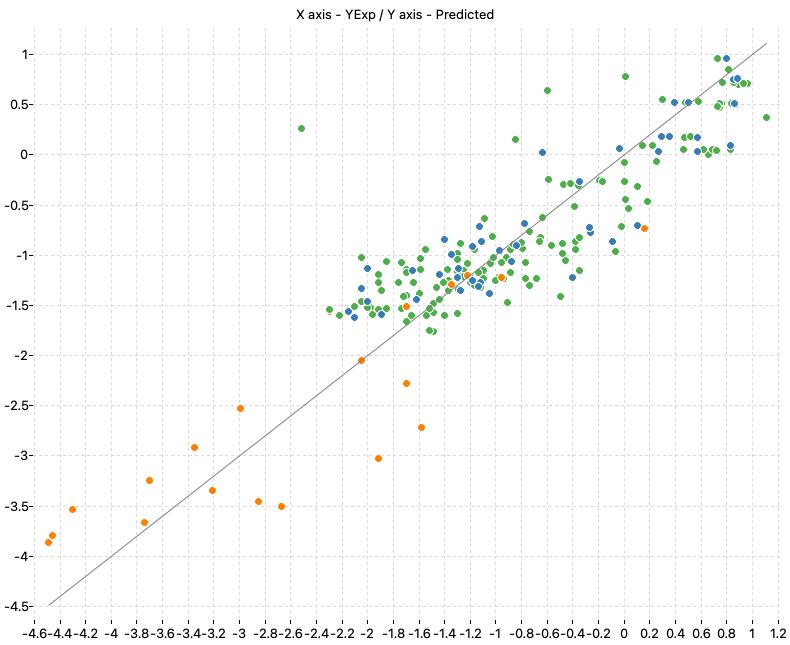
Prediction of acute toxicity towards the fathead minnow (Pimephales promelas)
This project includes the models presented in the paper “Cassotti, M., Ballabio, D., Todeschini, R., & Consonni, V. (2015). A similarity-based QSAR model for predicting acute toxicity towards the fathead minnow (Pimephales promelas). SAR and QSAR in Environmental Research, 26(3), 217–243. https://doi.org/10.1080/1062936X.2015.1018938“. The authors presented a study on the prediction of the acute toxicity of chemicals to fish. In particular, they presented QSAR models to predict the LC50 96 hours for the fathead minnow (Pimephales promelas).
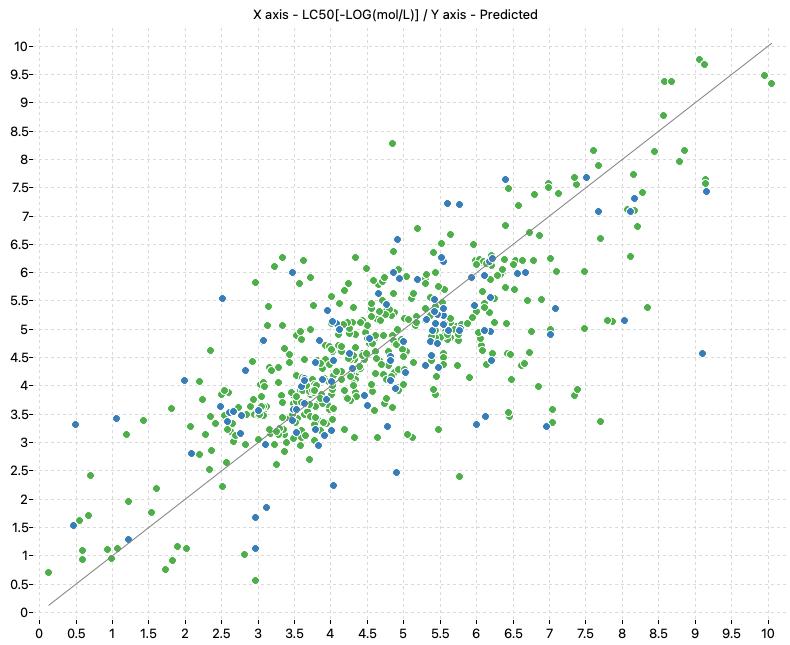
Prediction of acute toxicity towards the Daphnia magna
This project includes the models presented in the papers “Cassotti, M., Ballabio, D., Consonni, V., Mauri, A., Tetko, I. V., & Todeschini, R. (2014). Prediction of Acute Aquatic Toxicity toward Daphnia Magna by using the GA- k NN Method. Alternatives to Laboratory Animals, 42(1), 31–41. https://doi.org/10.1177/026119291404200106” and “Cassotti, M., Consonni, V., Mauri, A., & Ballabio, D. (2014). Validation and extension of a similarity-based approach for prediction of acute aquatic toxicity towards Daphnia magna. SAR and QSAR in Environmental Research, 25(12), 1013–1036. https://doi.org/10.1080/1062936X.2014.977818“. The authors presented QSAR models to predict acute aquatic toxicity (LC50 48 hours) towards Daphnia magna.
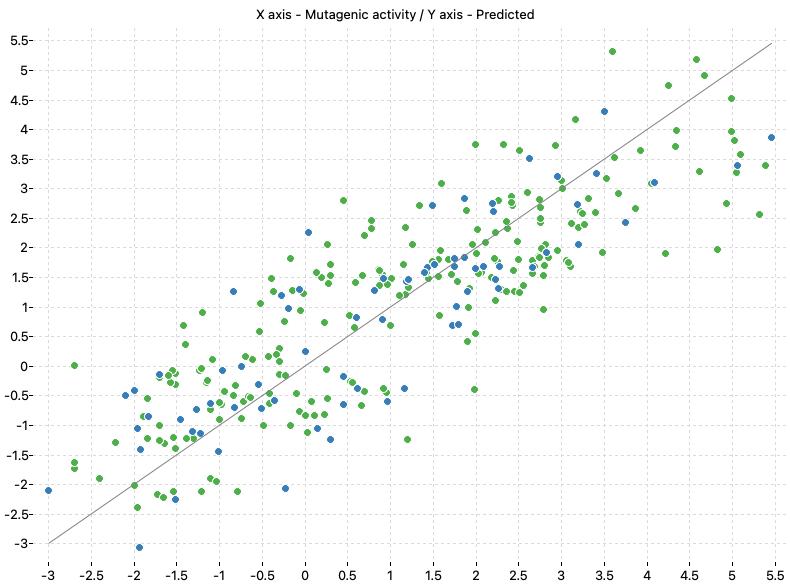
Prediction of mutagenicity of nitro and amino aromatic compounds against Salmonella typhimurium species
This project includes the models presented in the paper “Krishna, G., Khan, K., & Roy, K. (2020). Application of QSARs in identification of mutagenicity mechanisms of nitro and amino aromatic compounds against Salmonella typhimurium species. Toxicology in Vitro, 65(November 2019), 104768. https://doi.org/10.1016/j.tiv.2020.104768“. The authors presented QSAR models to predict mutagenicity mechanisms of nitro and amino aromatic compounds against Salmonella typhimurium species.
Prediction of ready biodegradability of chemicals
This project includes the models presented in the paper “Mansouri, K., Ringsted, T., Ballabio, D., Todeschini, R., & Consonni, V. (2013). Quantitative structure-activity relationship models for ready biodegradability of chemicals. Journal of Chemical Information and Modeling, 53(4), 867–878. https://doi.org/10.1021/ci4000213“. The authors presented four QSAR classification models to predict the ready biodegradability of chemicals.
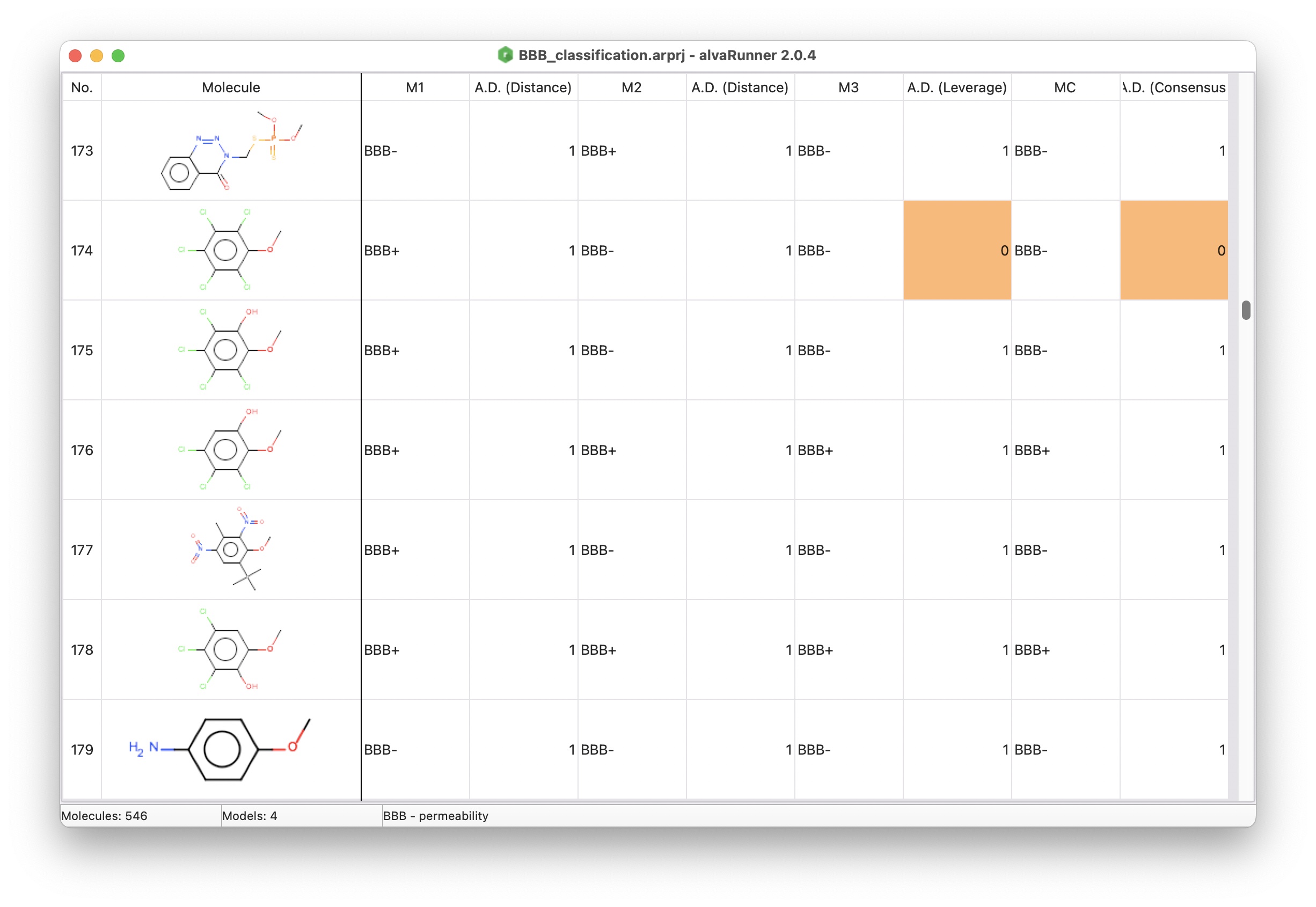
Prediction of Blood-Brain Barrier (BBB) permeability
This project includes the models described in the paper “Mauri, A., & Bertola, M. (2022). Alvascience : A New Software Suite for the QSAR Workflow Applied to the Blood – Brain Barrier Permeability. International Journal of Molecular Sciences, 23(21), 12882. https://doi.org/10.3390/ijms232112882“. The authors presented four QSAR classification models to predict the Blood-Brain Barrier (BBB) permeability.
Prediction of pesticides toxicity to earthworm
This project includes the model described in the paper “Ghosh, S., Ojha, P. K., Carnesecchi, E., Lombardo, A., Roy, K., & Benfenati, E. (2020). Exploring QSAR modeling of toxicity of chemicals on earthworm. Ecotoxicology and Environmental Safety, 190(December 2019), 110067. https://doi.org/10.1016/j.ecoenv.2019.110067“. The authors presented a study on the prediction of the toxicity of pesticides to earthworm.
Prediction of Bioconcentration Factor (BCF) of chemicals
This project includes a model described in the paper “Bhattacharyya, P., Samanta, P., Kumar, A., Das, S., & Ojha, P. K. (2024). Quantitative read-across structure–property relationship (q-RASPR): a novel approach to estimate the bioaccumulative potential for diverse classes of industrial chemicals in aquatic organisms. Environmental Science: Processes & Impacts. https://doi.org/10.1039/D4EM00374H“. The authors presented a study on the prediction of the Bioconcentration Factor (BCF) of chemicals.